Bayesian edge selection or Bayesian estimation for a Markov random field model for binary and/or ordinal variables.
Source:R/bgm.R
bgm.RdThe function bgm explores the joint pseudoposterior distribution of
parameters and possibly edge indicators for a Markov Random Field model for
mixed binary and ordinal variables.
Usage
bgm(
x,
variable_type = "ordinal",
reference_category,
iter = 10000,
burnin = 500,
interaction_scale = 2.5,
threshold_alpha = 0.5,
threshold_beta = 0.5,
edge_selection = TRUE,
edge_prior = c("Bernoulli", "Beta-Bernoulli", "Stochastic-Block"),
inclusion_probability = 0.5,
beta_bernoulli_alpha = 1,
beta_bernoulli_beta = 1,
dirichlet_alpha = 1,
lambda = 1,
na_action = c("listwise", "impute"),
save = FALSE,
display_progress = TRUE
)Arguments
- x
A data frame or matrix with
nrows andpcolumns containing binary and ordinal variables fornindependent observations andpvariables in the network. Regular binary and ordinal variables are recoded as non-negative integers(0, 1, ..., m)if not already done. Unobserved categories are collapsed into other categories after recoding (i.e., if category 1 is unobserved, the data are recoded from (0, 2) to (0, 1)). Blume-Capel ordinal variables are also coded as non-negative integers if not already done. However, since “distance” to the reference category plays an important role in this model, unobserved categories are not collapsed after recoding.- variable_type
What kind of variables are there in
x? Can be a single character string specifying the variable type of allpvariables at once or a vector of character strings of lengthpspecifying the type for each variable inxseparately. Currently, bgm supports “ordinal” and “blume-capel”. Binary variables are automatically treated as “ordinal’’. Defaults tovariable_type = "ordinal".- reference_category
The reference category in the Blume-Capel model. Should be an integer within the range of integer scores observed for the “blume-capel” variable. Can be a single number specifying the reference category for all Blume-Capel variables at once, or a vector of length
pwhere thei-th element contains the reference category for variableiif it is Blume-Capel, and bgm ignores its elements for other variable types. The value of the reference category is also recoded when bgm recodes the corresponding observations. Only required if there is at least one variable of type “blume-capel”.- iter
How many iterations should the Gibbs sampler run? The default of
1e4is for illustrative purposes. For stable estimates, it is recommended to run the Gibbs sampler for at least1e5iterations.- burnin
The number of iterations of the Gibbs sampler before saving its output. Since it may take some time for the Gibbs sampler to converge to the posterior distribution, it is recommended not to set this number too low. When
edge_selection = TRUE, the bgm function will perform2 * burniniterations, firstburniniterations without edge selection, thenburniniterations with edge selection. This helps ensure that the Markov chain used for estimation starts with good parameter values and that the adaptive MH proposals are properly calibrated.- interaction_scale
The scale of the Cauchy distribution that is used as a prior for the pairwise interaction parameters. Defaults to
2.5.- threshold_alpha, threshold_beta
The shape parameters of the beta-prime prior density for the threshold parameters. Must be positive values. If the two values are equal, the prior density is symmetric about zero. If
threshold_betais greater thanthreshold_alpha, the distribution is skewed to the left, and ifthreshold_betais less thanthreshold_alpha, it is skewed to the right. Smaller values tend to lead to more diffuse prior distributions.- edge_selection
Should the function perform Bayesian edge selection on the edges of the MRF in addition to estimating its parameters (
edge_selection = TRUE), or should it just estimate the parameters (edge_selection = FALSE)? The default isedge_selection = TRUE.- edge_prior
The inclusion or exclusion of individual edges in the network is modeled with binary indicator variables that capture the structure of the network. The argument
edge_prioris used to set a prior distribution for the edge indicator variables, i.e., the structure of the network. Currently, three options are implemented: The Bernoulli modeledge_prior = "Bernoulli"assumes that the probability that an edge between two variables is included is equal toinclusion_probabilityand independent of other edges or variables. Wheninclusion_probability = 0.5, this means that each possible network structure is given the same prior weight. The Beta-Bernoulli modeledge_prior = "Beta-Bernoulli"assumes a beta prior for the unknown inclusion probability with shape parametersbeta_bernoulli_alphaandbeta_bernoulli_beta. Ifbeta_bernoulli_alpha = 1andbeta_bernoulli_beta = 1, this means that networks with the same complexity (number of edges) get the same prior weight. The Stochastic Block modeledge_prior = "Stochastic-Block"assumes that nodes can be organized into blocks or clusters. In principle, the assignment of nodes to such clusters is unknown, and the model as implemented here considers all possible options (i.e., specifies a Dirichlet prior on the probability of allocations as described by Geng et al. 2019) . This model is advantageous when nodes are expected to fall into distinct clusters. The inclusion probabilities for the edges are defined at the level of the clusters, with a beta prior for the unknown inclusion probability with shape parametersbeta_bernoulli_alphaandbeta_bernoulli_beta, and a Dirichlet prior on the cluster assignment probabilities with a common concentration parameterdirichlet_alphaand a zero-truncated Poisson prior on the number of clusters with a rate parameterlambda, indicating the expected number of clusters. The default isedge_prior = "Bernoulli".- inclusion_probability
The prior edge inclusion probability for the Bernoulli model. Can be a single probability, or a matrix of
prows andpcolumns specifying an inclusion probability for each edge pair. The default isinclusion_probability = 0.5.- beta_bernoulli_alpha, beta_bernoulli_beta
The two shape parameters of the Beta prior density for the Bernoulli inclusion probability. Must be positive numbers. Defaults to
beta_bernoulli_alpha = 1andbeta_bernoulli_beta = 1.- dirichlet_alpha
The shape of the Dirichlet prior on the node-to-block allocation probabilities for the Stochastic Block model.
- lambda
The rate parameter of the zero-truncated Poisson prior on the number of cluster for the Stochastic Block model.
- na_action
How do you want the function to handle missing data? If
na_action = "listwise", listwise deletion is used. Ifna_action = "impute", missing data are imputed iteratively during the MCMC procedure. Since imputation of missing data can have a negative impact on the convergence speed of the MCMC procedure, it is recommended to run the MCMC for more iterations. Also, since the numerical routines that search for the mode of the posterior do not have an imputation option, the bgm function will automatically switch tointeraction_prior = "Cauchy"andadaptive = TRUE.- save
Should the function collect and return all samples from the Gibbs sampler (
save = TRUE)? Or should it only return the (model-averaged) posterior means (save = FALSE)? Defaults toFALSE.- display_progress
Should the function show a progress bar (
display_progress = TRUE)? Or not (display_progress = FALSE)? The default isTRUE.
Value
If save = FALSE (the default), the result is a list of class
“bgms” containing the following matrices with model-averaged quantities:
indicator: A matrix withprows andpcolumns, containing the posterior inclusion probabilities of individual edges.interactions: A matrix withprows andpcolumns, containing model-averaged posterior means of the pairwise associations.thresholds: A matrix withprows andmax(m)columns, containing model-averaged category thresholds. In the case of “blume-capel” variables, the first entry is the parameter for the linear effect and the second entry is the parameter for the quadratic effect, which models the offset to the reference category.In the case of
edge_prior = "Stochastic-Block", two additional elements are returned:A vector
allocationswith the estimated cluster assignments of the nodes, calculated using a method proposed by (Dahl2009) and also used by (GengEtAl_2019) .A matrix
componentswith the estimated posterior probability of the number of components (clusters) in the network. These probabilities are calculated based on Equation 3.7 in (miller2018mixture) , which computes the conditional probability of the number of components given the number of clusters. The number of clusters is derived from the cardinality of the sampledallocationsvector for each iteration of the MCMC sampler (seesave = TRUE).
If save = TRUE, the result is a list of class “bgms” containing:
indicator: A matrix withiterrows andp * (p - 1) / 2columns, containing the edge inclusion indicators from every iteration of the Gibbs sampler.interactions: A matrix withiterrows andp * (p - 1) / 2columns, containing parameter states from every iteration of the Gibbs sampler for the pairwise associations.thresholds: A matrix withiterrows andsum(m)columns, containing parameter states from every iteration of the Gibbs sampler for the category thresholds.In the case of
edge_prior = "Stochastic-Block"a matrixallocationswith the cluster assignments of the nodes from each iteration is returned. This matrix can be used to calculate the posterior probability of the number of clusters by utilizing thesummary_SBMfunction.
Column averages of these matrices provide the model-averaged posterior means.
Except for the allocations matrix, for which the summary_SBM
needs to be utilized.
In addition to the analysis results, the bgm output lists some of the arguments of its call. This is useful for post-processing the results.
Details
Currently, bgm supports two types of ordinal variables. The regular, default,
ordinal variable type has no restrictions on its distribution. Every response
category except the first receives its own threshold parameter. The
Blume-Capel ordinal variable assumes that there is a specific reference
category, such as the “neutral” in a Likert scale, and responses are scored
in terms of their distance to this reference category. Specifically, the
Blume-Capel model specifies the following quadratic model for the threshold
parameters:
$$\mu_{\text{c}} = \alpha \times \text{c} + \beta \times (\text{c} - \text{r})^2,$$
where \(\mu_{\text{c}}\) is the threshold for category c.
The parameter \(\alpha\) models a linear trend across categories,
such that \(\alpha > 0\) leads to an increasing number of
observations in higher response categories and \(\alpha <0\)
leads to a decreasing number of observations in higher response categories.
The parameter \(\beta\) models the response style in terms of an
offset with respect to the reference category \(r\); if \(\beta<0\)
there is a preference to respond in the reference category (i.e., the model
introduces a penalty for responding in a category further away from the
reference_category category r), while if \(\beta > 0\)
there is preference to score in the extreme categories further away from the
reference_category category.
The Bayesian estimation procedure (edge_selection = FALSE) simply
estimates the threshold and pairwise interaction parameters of the ordinal
MRF, while the Bayesian edge selection procedure
(edge_selection = TRUE) also models the probability that individual
edges should be included or excluded from the model. Bayesian edge selection
imposes a discrete spike and slab prior distribution on the pairwise
interactions. By formulating it as a mixture of mutually singular
distributions, the function can use a combination of Metropolis-Hastings and
Gibbs sampling to create a Markov chain that has the joint posterior
distribution as an invariant. The current option for the slab distribution is
a Cauchy with an optional scaling parameter. The slab distribution is also used
as the prior for the interaction parameters for Bayesian estimation. A
beta-prime distribution is used for the exponent of the category parameters.
For Bayesian edge selection, two prior distributions are implemented for the
edge inclusion variables (i.e., the prior probability that an edge is
included); the Bernoulli prior and the Beta-Bernoulli prior.
References
Geng J, Bhattacharya A, Pati D (2019). “Probabilistic community detection with unknown number of communities.” Journal of the American Statistical Association, 114, 893–905. doi:10.1080/01621459.2018.1458618 .
Examples
# \donttest{
#Store user par() settings
op <- par(no.readonly = TRUE)
##Analyse the Wenchuan dataset
# Here, we use 1e4 iterations, for an actual analysis please use at least
# 1e5 iterations.
fit = bgm(x = Wenchuan)
#> Warning: There were 18 rows with missing observations in the input matrix x.
#> Since na_action = listwise these rows were excluded from the analysis.
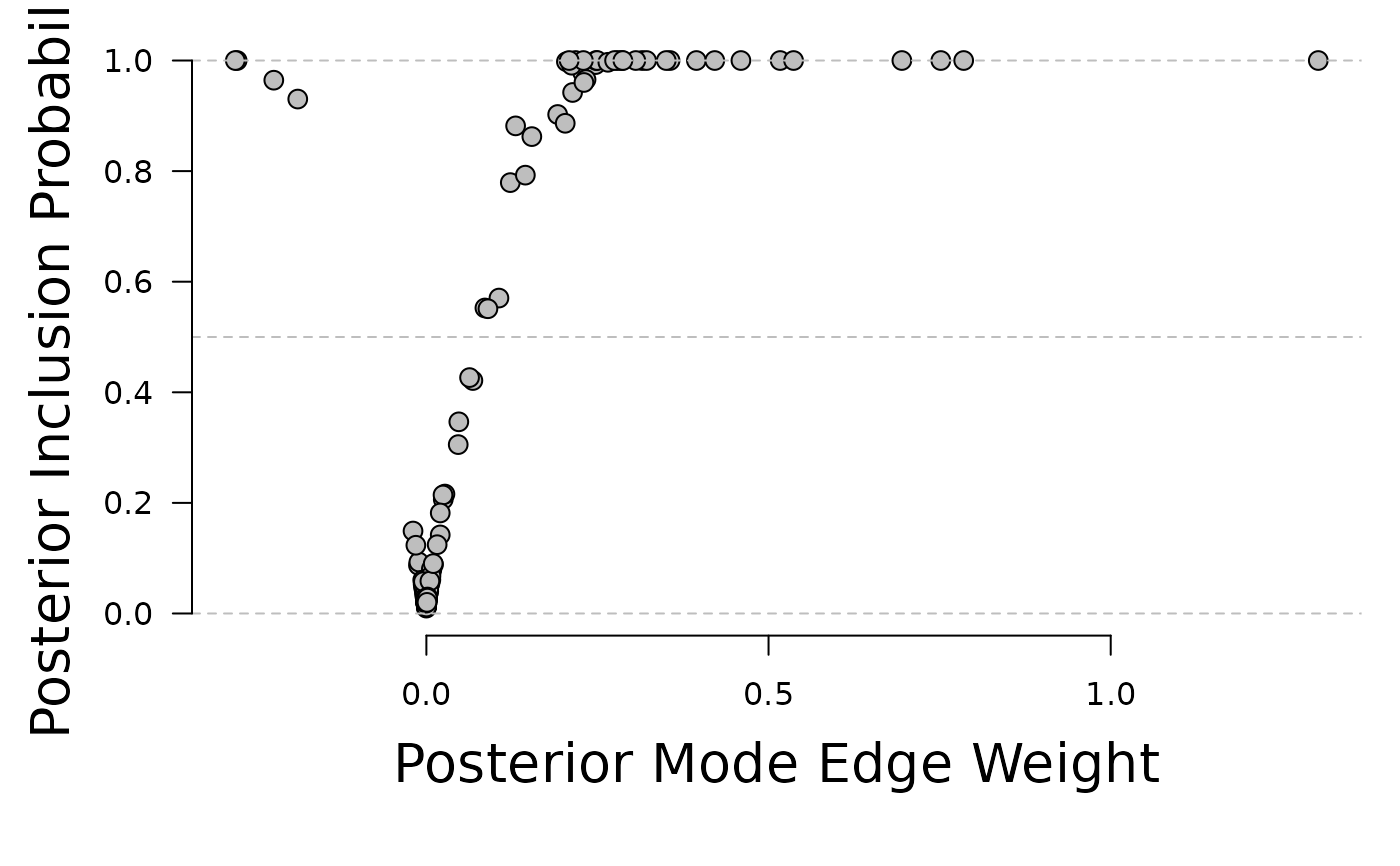
#------------------------------------------------------------------------------|
# INCLUSION - EDGE WEIGHT PLOT
#------------------------------------------------------------------------------|
par(mar = c(6, 5, 1, 1))
plot(x = fit$interactions[lower.tri(fit$interactions)],
y = fit$indicator[lower.tri(fit$indicator)], ylim = c(0, 1),
xlab = "", ylab = "", axes = FALSE, pch = 21, bg = "gray", cex = 1.3)
abline(h = 0, lty = 2, col = "gray")
abline(h = 1, lty = 2, col = "gray")
abline(h = .5, lty = 2, col = "gray")
mtext("Posterior Mode Edge Weight", side = 1, line = 3, cex = 1.7)
mtext("Posterior Inclusion Probability", side = 2, line = 3, cex = 1.7)
axis(1)
axis(2, las = 1)
 #------------------------------------------------------------------------------|
# EVIDENCE - EDGE WEIGHT PLOT
#------------------------------------------------------------------------------|
#For the default choice of the structure prior, the prior odds equal one:
prior.odds = 1
posterior.inclusion = fit$indicator[lower.tri(fit$indicator)]
posterior.odds = posterior.inclusion / (1 - posterior.inclusion)
log.bayesfactor = log(posterior.odds / prior.odds)
log.bayesfactor[log.bayesfactor > 5] = 5
par(mar = c(5, 5, 1, 1) + 0.1)
plot(fit$interactions[lower.tri(fit$interactions)], log.bayesfactor, pch = 21, bg = "#bfbfbf",
cex = 1.3, axes = FALSE, xlab = "", ylab = "", ylim = c(-5, 5.5),
xlim = c(-0.5, 1.5))
axis(1)
axis(2, las = 1)
abline(h = log(1/10), lwd = 2, col = "#bfbfbf")
abline(h = log(10), lwd = 2, col = "#bfbfbf")
text(x = 1, y = log(1 / 10), labels = "Evidence for Exclusion", pos = 1,
cex = 1.7)
text(x = 1, y = log(10), labels = "Evidence for Inclusion", pos = 3, cex = 1.7)
text(x = 1, y = 0, labels = "Absence of Evidence", cex = 1.7)
mtext("Log-Inclusion Bayes Factor", side = 2, line = 3, cex = 1.5, las = 0)
mtext("Posterior Mean Interactions ", side = 1, line = 3.7, cex = 1.5, las = 0)
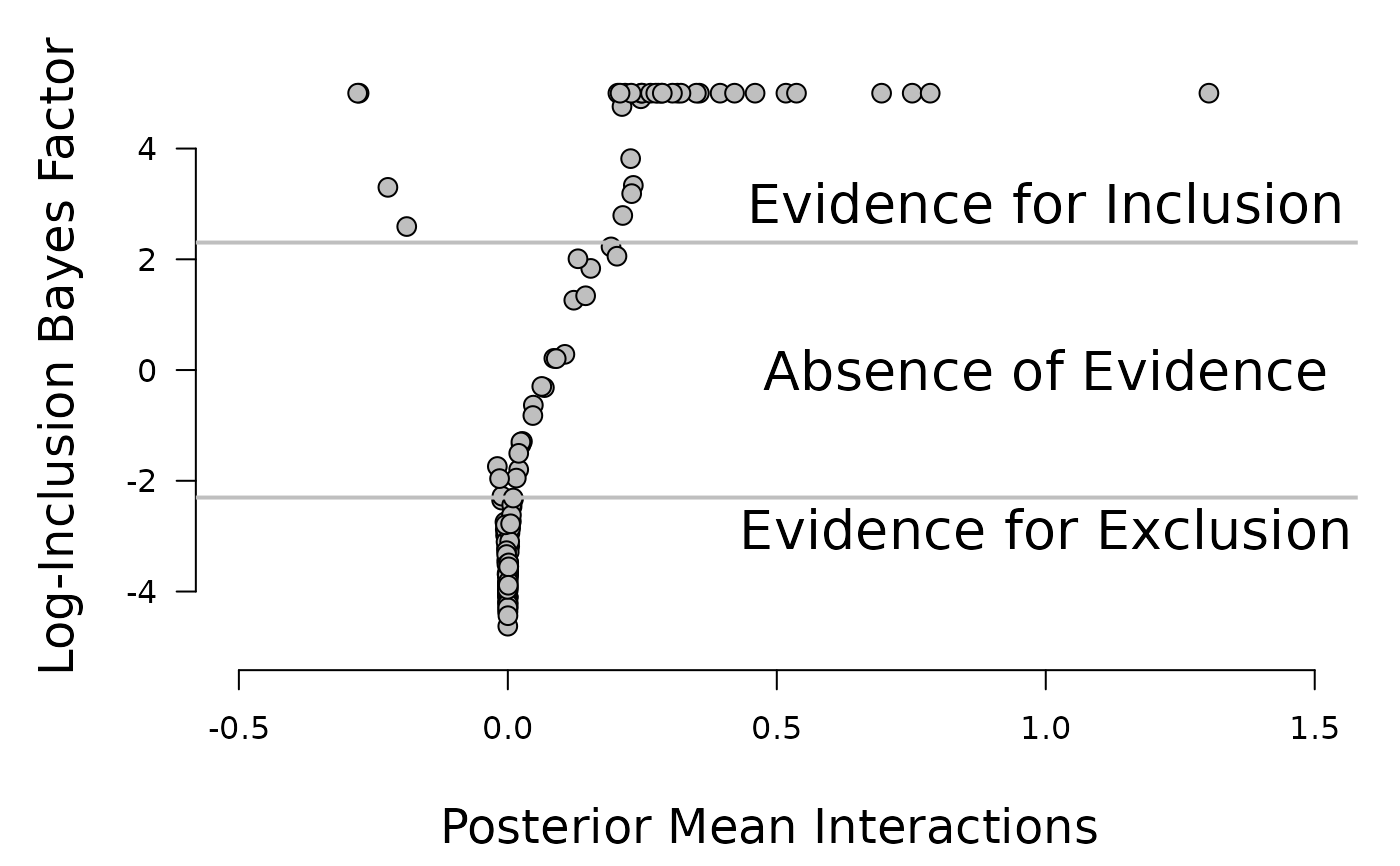
#------------------------------------------------------------------------------|
# EVIDENCE - EDGE WEIGHT PLOT
#------------------------------------------------------------------------------|
#For the default choice of the structure prior, the prior odds equal one:
prior.odds = 1
posterior.inclusion = fit$indicator[lower.tri(fit$indicator)]
posterior.odds = posterior.inclusion / (1 - posterior.inclusion)
log.bayesfactor = log(posterior.odds / prior.odds)
log.bayesfactor[log.bayesfactor > 5] = 5
par(mar = c(5, 5, 1, 1) + 0.1)
plot(fit$interactions[lower.tri(fit$interactions)], log.bayesfactor, pch = 21, bg = "#bfbfbf",
cex = 1.3, axes = FALSE, xlab = "", ylab = "", ylim = c(-5, 5.5),
xlim = c(-0.5, 1.5))
axis(1)
axis(2, las = 1)
abline(h = log(1/10), lwd = 2, col = "#bfbfbf")
abline(h = log(10), lwd = 2, col = "#bfbfbf")
text(x = 1, y = log(1 / 10), labels = "Evidence for Exclusion", pos = 1,
cex = 1.7)
text(x = 1, y = log(10), labels = "Evidence for Inclusion", pos = 3, cex = 1.7)
text(x = 1, y = 0, labels = "Absence of Evidence", cex = 1.7)
mtext("Log-Inclusion Bayes Factor", side = 2, line = 3, cex = 1.5, las = 0)
mtext("Posterior Mean Interactions ", side = 1, line = 3.7, cex = 1.5, las = 0)
 #------------------------------------------------------------------------------|
# THE MEDIAN PROBABILITY NETWORK
#------------------------------------------------------------------------------|
tmp = fit$interactions[lower.tri(fit$interactions)]
tmp[posterior.inclusion < 0.5] = 0
median.prob.model = matrix(0, nrow = ncol(Wenchuan), ncol = ncol(Wenchuan))
median.prob.model[lower.tri(median.prob.model)] = tmp
median.prob.model = median.prob.model + t(median.prob.model)
rownames(median.prob.model) = colnames(Wenchuan)
colnames(median.prob.model) = colnames(Wenchuan)
library(qgraph)
qgraph(median.prob.model,
theme = "TeamFortress",
maximum = .5,
fade = FALSE,
color = c("#f0ae0e"), vsize = 10, repulsion = .9,
label.cex = 1.1, label.scale = "FALSE",
labels = colnames(Wenchuan))
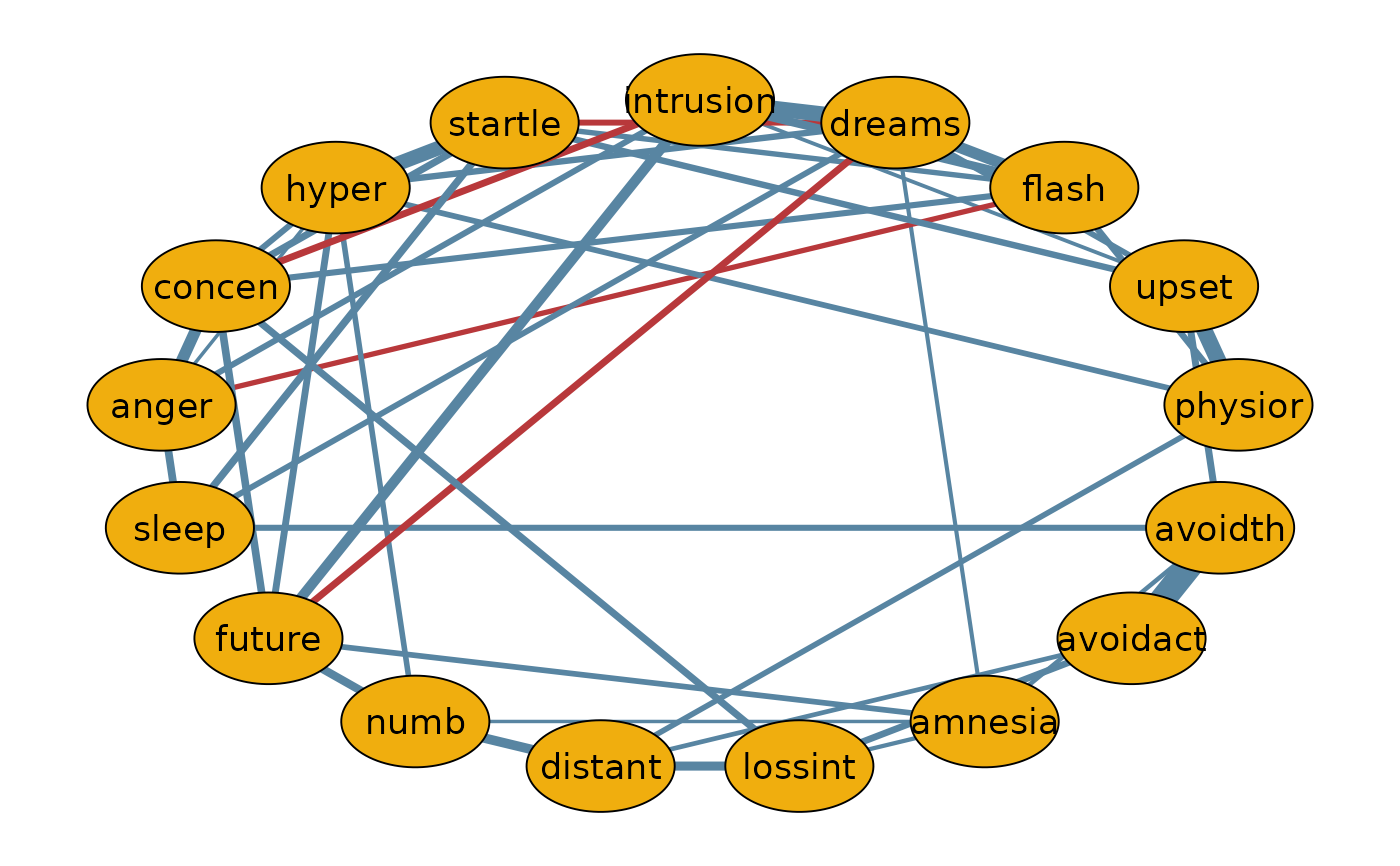
#------------------------------------------------------------------------------|
# THE MEDIAN PROBABILITY NETWORK
#------------------------------------------------------------------------------|
tmp = fit$interactions[lower.tri(fit$interactions)]
tmp[posterior.inclusion < 0.5] = 0
median.prob.model = matrix(0, nrow = ncol(Wenchuan), ncol = ncol(Wenchuan))
median.prob.model[lower.tri(median.prob.model)] = tmp
median.prob.model = median.prob.model + t(median.prob.model)
rownames(median.prob.model) = colnames(Wenchuan)
colnames(median.prob.model) = colnames(Wenchuan)
library(qgraph)
qgraph(median.prob.model,
theme = "TeamFortress",
maximum = .5,
fade = FALSE,
color = c("#f0ae0e"), vsize = 10, repulsion = .9,
label.cex = 1.1, label.scale = "FALSE",
labels = colnames(Wenchuan))
 #Restore user par() settings
par(op)
# }
#Restore user par() settings
par(op)
# }